Hi, I'm Lingli
Research & Development Professional
Passionate about utilizing the machine learning technique and developing computational workflow to solve Cryo-EM challenges and always eager to collaborate on innovative projects.

Passionate about utilizing the machine learning technique and developing computational workflow to solve Cryo-EM challenges and always eager to collaborate on innovative projects.

I’m a computational scientist building physics-aware algorithms and AI tools that turn messy, in-situ cryo-EM data into reliable structure. I’ve contributed multiple C++ programs to the cisTEM suite and related tools (e.g., Unbend for local motion correction, CTFFIND5 tilt components, fit_tilt_model, shift_field_generation), and I develop an orientation-conditioned neural network that augments 2D template matching (2DTM) for membrane-protein detection from liposomes—reducing template bias and enabling one-step reconstructions with minimal additional refinement. My tools are delivered as production-ready C++/Python with reproducible benchmarks. Current focus: a generalizable machine learning model for membrane protein detection from in situ data and calibrated end-to-end pipelines that reduce manual tuning and accelerate 3D reconstructions.
Advanced computational methods for processing cryo-electron microscopy images, focusing on motion correction and contrast transfer function estimation.
Learn MoreApplying machine learning and deep learning techniques to solve complex problems in cryo-EM data processing and structural analysis.
Learn MoreDeveloping robust computational tools and software packages that advance the field of structural biology and cryo-EM.
Learn More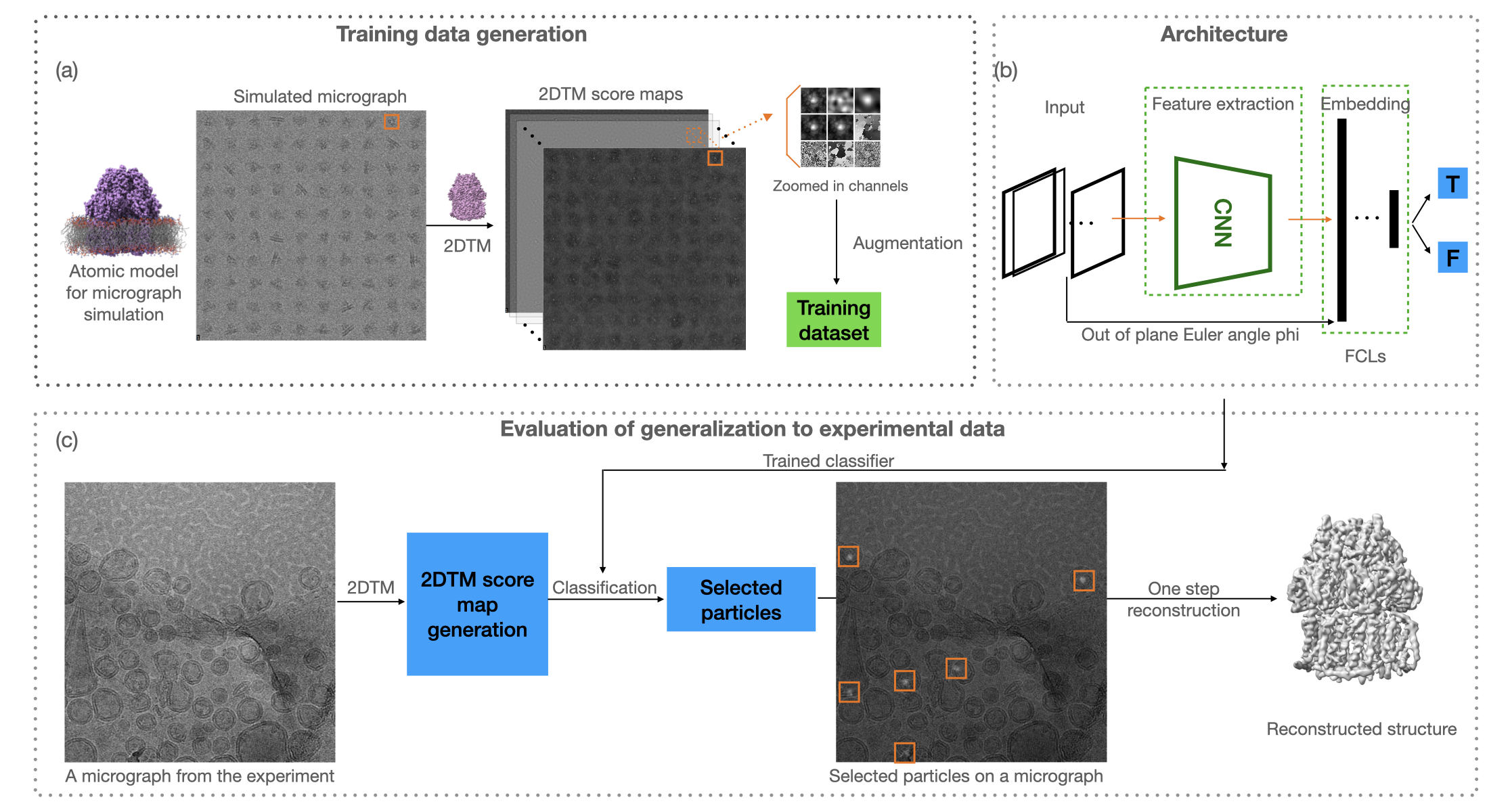
This project focuses on the detection of membrane proteins from liposome structures using advanced deep learning techniques. Key achievements include the development of a novel 2D template matching algorithm and the successful application of convolutional neural networks for feature extraction.
This project focuses on correcting local motion artifacts in cryo-electron microscopy (Cryo-EM) movie frames using advanced image processing techniques. Key achievements include the development of a novel algorithm for motion correction and the successful application of deep learning for artifact removal.
A CNN-BiLSTM that learns local edge shapes and longer-range spectral context, trained on simulated + curated experimental spectra.
Electron Energy Loss Spectroscopy (EELS) is powerful but slow and operator-dependent: background subtraction, low SNR, and overlapping edges make peak identification error-prone—especially in high-throughput EM workflows.
Loading publications...
I'm actively seeking new opportunities and would love to discuss how my research and development experience that can contribute to your team.